Once upon a time, there was simplicity (or at least perceived simplicity) in the way that cells communicated with each other. There were proteins (e.g., growth factors) and non-proteins (e.g., steroid hormones) passing from one cell to another in a never-ending conversation. Then miRNA became the star of the show (as did everyone’s publications), complicating cell-cell communication. Not too long after, extracellular vesicles (EVs) broke into the zeitgeist, with an ever-expanding list of subtypes. EVs have reigned for some time as the ‘it’ particles, but now there are new kids on the cell-cell communication block. Two smaller, more agile models are vying for the attention of researchers. Meet the exomeres and the supermeres.
What are exomeres and supermeres?
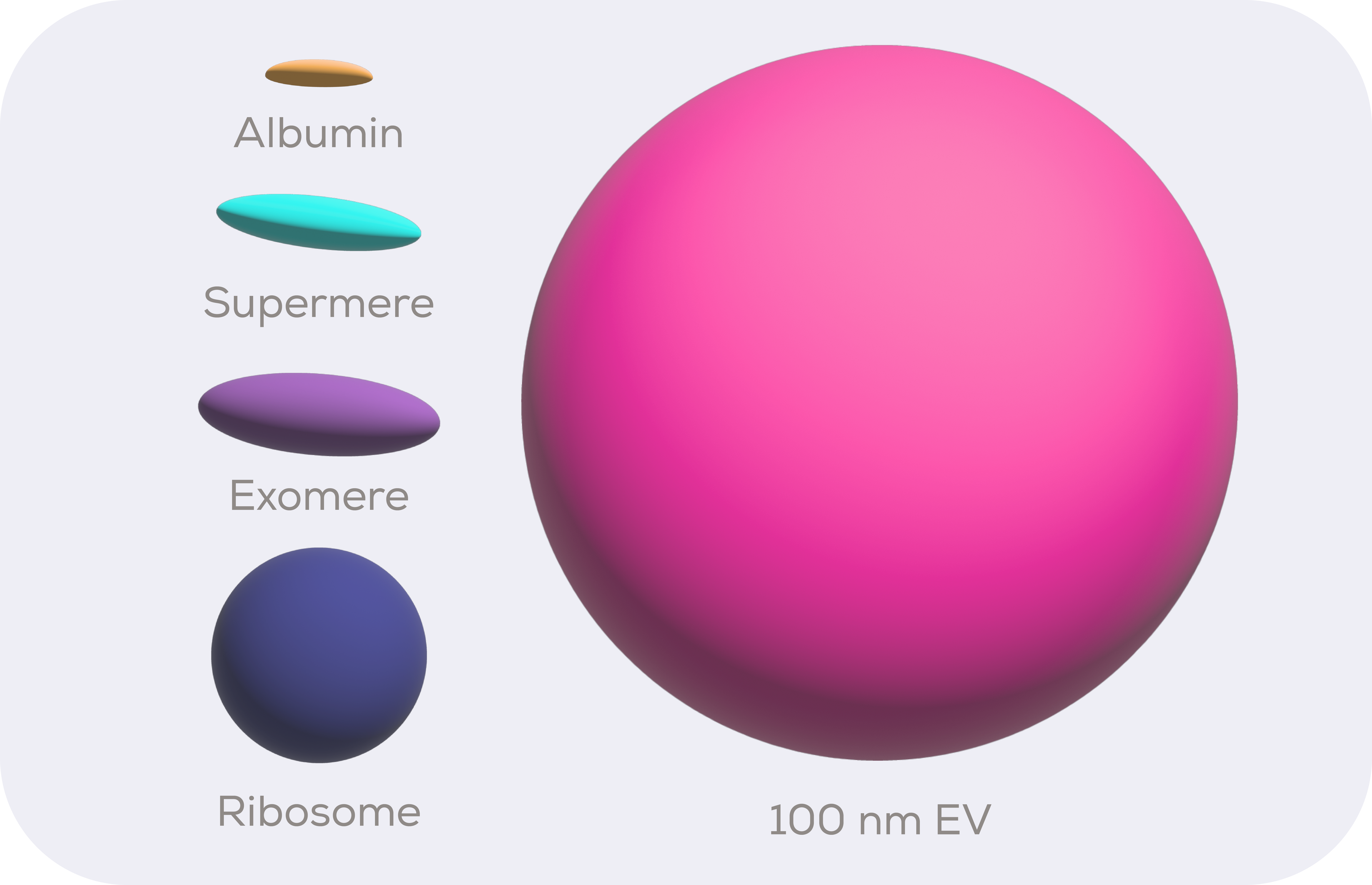
Despite being somewhat of a highlight at EV conferences over the last few years, exomeres are in fact non-vesicular and so not EVs. Instead, they are mostly protein-based. They are also quite a bit smaller than EVs, being around 11 x 34 nm in size as determined by fluid phase atomic force microscopy (AFM).1 Assuming an ellipsoid morphology, this makes them around 80 times smaller than a hypothetical 100 nm diameter small EV (sEV).
Once everything exomere-size and above have been stripped from a solution, the supermeres – or supernatant of exomeres – are the next biggest things. They are around 7 x 29 nm in size as determined by AFM, which puts them at just under half the size of exomeres.1 Which, not to put too fine a point on it, is absolutely tiny. The protein albumin is less than 8 times smaller than a supermere and by volume ribosomes are bigger then exomeres, which does raise the question of when something becomes a particle rather than just a protein complex/aggregate.
How accurate are the methods used to analyse exomere and supermere size?
For particle-by-particle techniques, only transmission electron microscopy and AFM are currently capable of accurately measuring the size of exomeres and supermeres. The major drawbacks of these methods are the long processing time and the low number of particles analysed. Seeing higher throughput single particle measurement technologies develop to meet the needs of the supermere and exomere field is essential for its progression.
What are exomeres made of and what do they do?
The biogenesis and composition of exomeres is currently mostly unknown. However, we do know that the galectin-3 binding protein (LGALS3BP) is abundant in exomeres and, conveniently, self-assembles into doughnut shaped decamers around the size of exomeres and might facilitate interaction with cells.2 This all offers a perhaps too tempting opportunity to believe that LGALS3BP proteins make up the skeleton or even the bulk of exomeres, but this is likely an oversimplification. We know that a plethora of other proteins – including ribosomal proteins and even the ectodomain of the SARS-CoV-2 receptor ACE2 – are present in exomeres.3 Despite this, research into their functions is sparse on the ground and rather inconclusive to date. Supermere research has had rather more luck.
What are supermeres made of and what do they do?
Supermeres are internalised into cells by clathrin-mediated endocytosis and micropinocytosis1, both of which also employed by sEVs entering cells.4 In vivo, supermeres are internalised into the regular detoxification/excretion organs but also into the lung and heart.1 Even more impressively, their tiny size allows them to cross the blood-brain barrier.
For a particle able to pass in and out of the blood-brain barrier, it is perhaps particularly interesting that supermeres contain the shed ectodomains of proteins such as amyloid precursor protein which are associated with Alzheimer’s disease.1 They are also enriched in glycolytic proteins such as enolase proteins which, among other functions, increase lactate concentrations. Zhang et al (2021) identified a potential role for supermeres in transferring cetuximab resistance from resistant to sensitive cells, possibly via a lactate induction mechanism.1 The same study found that supermeres impacted upon hepatic lipid and glycogen handling in mice, perhaps through modulation of mTORC1 signalling. It is clear then that whatever they are, supermeres are functional.
Extracellular RNA in supermeres and exomeres
The type 2 diabetes associated miRNA, miR-1246, is 1,024-fold enriched in supermeres vs cells.1 Interestingly, this miRNA is not necessarily spliced in cells, but is a product of the degradation of the U2 small nuclear RNA (snRNA)3 by extracellular exonucleases. The portion of U2 snRNA which corresponds to the sequence of miR-1246 is protected from degradation in supermeres by proteins of the pre-catalytic splicosome.3 This supports the Tosar et al. (2021) theory that “survivorship defines the extracellular RNA landscape”5, with miRNAs in the extracellular environment needing to be protected to survive or even to come into existence.
Whilst supermeres and exomeres have distinct small RNA profiles6, in part due to the high number of RNA-binding proteins in supermeres1, having distinct content and being functional doesn’t necessarily mean that they are produced or even present in vivo.
Could isolation method shape exomeres and supermeres?
Exomeres and supermeres are diverse1, which is evident from their proteomics which consists of more proteins than you would expect to fit in their tiny size. This raises the question of whether the categories exomeres and supermere truly exist as distinctly produced particles, or whether they simply represent size classes of protein complexes or even aggregates induced by the extreme ultracentrifugation forces employed to isolate them.
Following the removal of EVs by slower ultracentrifugation speeds (e.g., 100,000 x g), exomeres are isolated from the 100,000 x g supernatant by an ultracentrifugation pelleting speed of 167,000 x g1. Supermeres are then pelleted from the 167,000 x g supernatant at a pelleting speed of 367,000 x g.1 EV aggregation occurs from as little as 100,000 x g11, much gentler than the speeds used to isolate exomeres and supermeres. As such, the use of gentler methods, such as size exclusion chromatography, and greater emphasis on identifying biogenesis routes must be prioritised in exomeres and supermere research.
Could extracellular vesicle isolates from ultracentrifugation contain exomeres and supermeres?
Our analysis of data from EV-TRACK shows that over 30% of EVs studies pellet EVs at forces above 100,000 x g and over 3% of studies pellet EVs at speeds at or above the exomeres’ isolation speed of 167,000 x g. Now that we know about the existence of exomeres, this raises the question of whether exomeres – perhaps even supermeres? – could contaminate high speed UC EV isolates.
As would be expected, we found that pelleting g force for EV isolation was negatively correlated with particle size in data reported by authors in EV-TRACK (Pearson r = -0.32, p < 0.00001). Indeed, LGALS3BP, a marker of exomeres, is highly abundant in the proteomics of EV isolates pelleted at or above 167,000 x g.7-10 It is also the 77th most identified protein in EV isolates according to Vesiclepedia. Enolase 1, a potential supermere marker, is the 9th most identified protein in EVs according to Vesiclepedia. Whilst this is far from definitive, it does suggest that further research in this area is needed.
The future of exomeres and supermere research
The most important thing for the exomere and supermere fields is to keep developing. They are both very new and we don’t know enough about them to be able to really, confidently say what they are or where they come from. One of the most important things will be to draw a line in the sand between what is a protein complex and what is a particle. Where do exomeres and supermeres fall in relation to this line? Are they just a size category of something we already knew about such as protein complexes? This field is likely to develop quickly, but to do so effectively, technology needs to catch up with the curiosity of the scientists pushing this research forwards.
Looking to isolate exomeres and supermeres in your research? Learn about the 20 nm series here









