What are extracellular vesicles?
Extracellular vesicles (EVs) are nanometre-scale, cell-derived ‘packages’ surrounded by a lipid bilayer. Parcelled within are a complex variety of proteins, RNAs1 (e.g. microRNAs), DNA2 and small molecules, all protected from degradation in the extracellular environment by the surrounding membrane. This membrane itself contains proteins and even a ‘decoration’ of nucleic acids. These EV ‘packages’ are secreted by all known cells, from bacteria to B cells, and are present in the full range of biofluids. Given their dynamic plasticity in response to the cellular environment, disease state and cell type, these finely tuned EVs contain a wealth of information ready for discovery in basic biological research, and in the development of EV-based diagnostics and therapeutics.
Whilst mighty in terms of their potential contribution to science and medicine, there is one inconvenient truth: EVs inherently reside in complex samples. Alongside EVs, biofluids and conditioned media contain a variety of biomolecules including proteins, nucleic acids, lipids, small molecules and lipoproteins. This presents challenges for isolating EVs in a way that retains EV composition, structure and functionality, whilst preventing the co-isolation of unwanted contaminants.
To address this, a variety of isolation methods have been developed in this rapidly evolving field.
The landscape of EV isolation
Initial isolation methods relied solely on ultracentrifugation (UC), where EVs were pelleted using extreme gravitational force. Differential UC uses initial less extreme centrifugation steps to pellet cells and larger debris, before a final high gravitational force step(s) is used to pellet EVs from the remaining supernatant. Further refinement using density gradients have since been implemented into UC workflows to improve the resolution of separation.
Precipitation was another early method. Polymers such as polyethylene glycol (known as PEG) are used to form a mesh within which EVs are trapped, allowing them to be pelleted, along with the polymer, at substantially lower gravitational forces. However, this method also traps non-EV components such as protein, making it non-specific3. Also, whilst faster and gentler than UC methods, the presence of the polymer complexed with EVs may hinder downstream applications.
Affinity-based ‘pull-downs’ based on known surface markers (e.g., proteins or lipids) have been developed; they rely on a certain degree of prior understanding of the composition of EVs in the sample and will only isolate subsets of EVs.
Finally, techniques based upon size have also been developed and adapted for EV studies, including filtration, flow field-flow fractionation, and size exclusion chromatography (SEC). Although UC remains a common form of EV isolation, the last few years have seen a rapid proliferation of the use of SEC4. This switch away from UC is likely to continue, as studies which began prior to commercially available SEC columns give way to studies arising in the modern era of EV isolation.
Here at Izon, we have developed SEC-based columns to isolate EVs from a diverse range of biological sources. qEV columns are available in different sizes to support a wide range of sample loading volumes from 150 µL to 100 mL, recently made available in the ‘Gen 2’ range, using a high perfoming, customised resin which delivers even purer EVs than the existing Legacy column range.
EV isolation by SEC maximises purity
The question of purity in EV isolation is an important one as the co-isolation of non-EV components may impact upon downstream applications. The most commonly co-isolated contaminants are proteins and lipoproteins.
A 2021 analysis of commercial EV isolation techniques by Veerman et al5 offers an insight into how well SEC performs in terms of EV purity compared to other techniques. qEV column isolates from both plasma and conditioned cell culture media contained the most known EV proteins of the studied techniques5. Whilst an affinity method also performed well in this respect, EDTA incompatibility and low particle concentration makes this method entirely unsuitable for plasma isolations and poor for other sample types5. Furthermore, commercial affinity-based products have been shown to co-isolate contaminate proteins, including albumin, to a far greater degree than qEV columns.6 SEC also has a significantly higher particle-to-protein ratio than UC7, reinforcing its superiority in terms of purity. In-house data (Figure 1) shows the ability of Gen 2 qEV columns to separate EVs from protein.
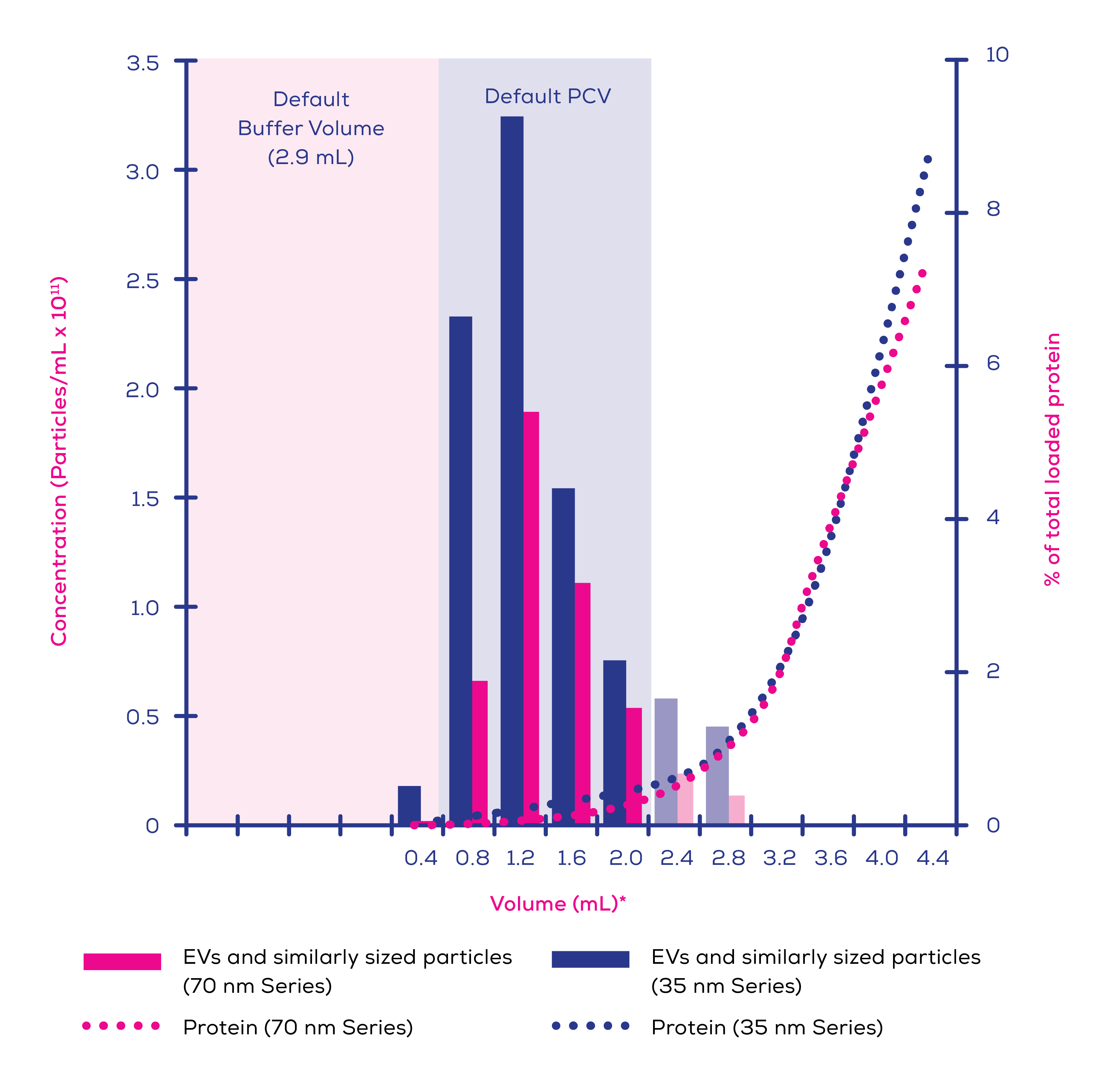
Figure 1. Eluted protein and extracellular vesicles (EVs) and similarly sized particles >60 nm in human plasma (0.5 mL loading volume) separated on qEVoriginal/35 nm Gen 2 and qEVoriginal/70 nm Gen 2 columns. EV concentration was measured using an Exoid and protein levels by bicinchoninic acid (BCA) assay. Faded bars represent calculated individual concentrations based on pooled sample measurements.*Volumes are labelled as the highest volume in that sample i.e., label ‘0.4’ refers to the volume from 0.0-0.4 mL after the buffer volume, label ‘0.8’ refers to the volume from 0.4-0.8 mL after the buffer volume, etc.
In comparison to soluble proteins, lipoproteins prove more complex to remove for all methods. Separating by density (including UC) leads to the co-isolation of EV density-similar lipoproteins (i.e. high-density lipoproteins)8, whilst separating by size (e.g. by SEC) results in the co-isolation of larger lipoproteins (i.e. very low-density lipoproteins and chylomicrons) with size ranges which overlap with that of EVs. This makes it all but impossible to entirely remove lipoproteins with a single EV isolation method.
For applications where depletion of lipoproteins is critical (e.g., EVs as therapeutics derived from lipoprotein-producing cells), it may be necessary to combine techniques to achieve the purest possible samples. However, a two-step process will not be necessary for many applications, considering most cells do not produce lipoproteins in substantial quantities. As such, your cell culture cell of choice may not produce meaningful levels of contaminating lipoproteins (Figure 2). Meanwhile, plasma is rich in lipoproteins and will require a two-step process if lipoprotein removal is a high priority. For example, cation exchange chromatography9, density gradient UC3 and ultrafiltration10 have been shown to pair with isolation by SEC to provide lipoprotein-depleted samples. Therefore, whilst no single method can completely deplete lipoproteins, SEC is the method of choice for inclusion in two-step processes.
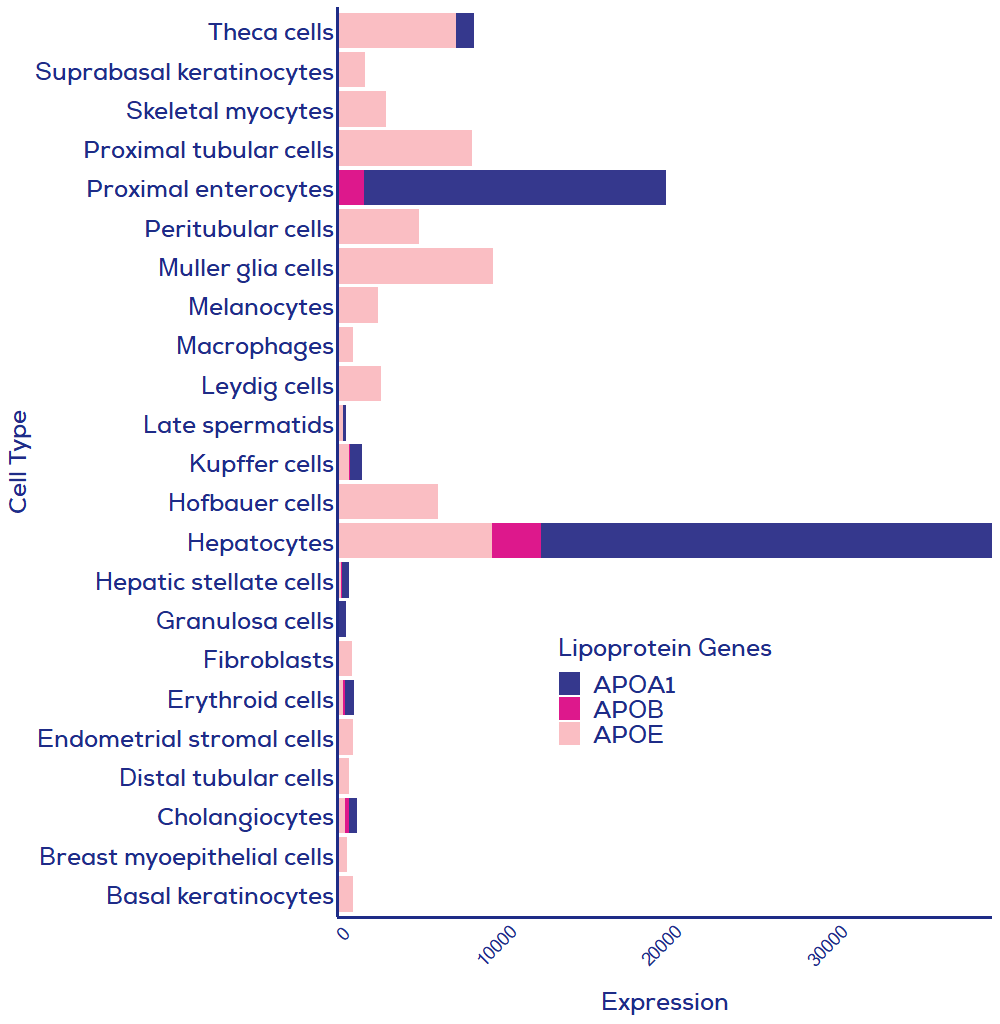
Figure 2. Human cell expression of three key lipoprotein genes. Data from single cell analysis of human cells where summed relative expression of the three presented lipoprotein genes was greater than 500. All cell types were included in the single cell RNAseq dataset, meaning that if your cell of interest is missing then expression of lipoproteins was lower than in cells included in this graph. APOA1 = HDL; APOB = chylomicrons, LDL, VLDL; APOE = chylomicrons remnants, VLDL, LDL, IDL and HDL. Data credit: Human Protein Atlas11
Reproducibility a key focus of qEV isolation
Reproducibility is essential in the push for standardisation in EV research. The minimal information for studies on extracellular vesicles (MISEV)12 is a semi-regularly updated position paper from the wider EV community where minimal standards for research and reporting are proposed. To improve reproducibility between studies, EV-TRACK is used to record the methodology of participating EV studies, allowing for maximal reproducibility. However, reproducibility is limited by the capabilities of the isolation methods used.
UC has poor reproducibility between laboratories, even when the same starting sample and UC protocol are used.13 Given the reliance of density-based isolation methods on UC, they are likely to suffer the same lack of reproducibility as UC alone. Due to the wide variety of ultracentrifuges and UC rotors in use, as well as variation introduced by users and differences in sample viscosity, it is unlikely that variability could be adequately removed from UC.
Commercial products can improve reproducibility by reducing the inter-user variation in ‘setting up’ the technique (e.g., preparation of gradients or SEC columns). qEV columns in themselves solve this problem by providing users with an optimised and standardised isolation column. However, standardised SEC columns cannot alone remove the impact of how the user performs the technique; user variation in the exact volume of each collected fraction could, amongst other steps, introduce variation into the method. This is where the Automatic Fraction Collector (AFC) steps in to reduce this user-introduced variation and improve reproducibility. By precisely weighing the fractions as they are collected, purified collection volumes (PCV) are standardised between isolations.
To provide flexibility, the PCV can be programmed by the user to suit individual sample types and downstream applications. Column cleaning is also prompted by the AFC, providing you with the opportunity to ensure qEV columns are in a standardised state after each use; this is beneficial for those who want to minimise residual sample carryover. Finally, the AFC also keeps track of the number of uses of each column to help avoid use beyond the column’s optimum lifespan.
The AFC and qEV pairing provides excellent reproducibility and together, they ensure that an exact and reproducible programme of isolation can be performed on every sample both within and between laboratories.
SEC is a gentle isolation method
Whilst UC remains a commonly used EV isolation method, UC subjects EVs to intense gravitational force. This results in EV degradation, aggregation and fusion, which does not occur with SEC.14,15 SEC is also gentler than techniques where pores or channels are used to separate by size (e.g., filtration), where blocking of the pores/channels by EVs and/or protein can result in shear stress which can potentially damage EVs.16,17 The absence of any applied force makes SEC the gentlest of techniques, allowing EVs to be isolated in the closest state possible to that in which they appeared in the starting material. The short processing time (around 15 minutes/sample) of qEV columns also means that EVs are not subjected to unfavourable temperature conditions for long periods of time, preserving their integrity.
Unsurprisingly, gentle isolation of EVs using SEC results in greater functionality of SEC-isolated EVs as compared with UC-isolated EVs.15,18 Fluorescently labelled EVs isolated using SEC also have different and more specific biodistribution than UC-isolated EVs when injected into mice.18 EVs isolated from cardiac progenitor cells phosphorylate the signalling kinase ERK in microvascular endothelial cells to a greater degree when isolated using SEC than with UC.15 Another gentle approach is precipitation; however, co-culture of THP-1 cells or macrophages with THP-1 EVs isolated using precipitation methods resulted in cell death, whilst EVs isolated using SEC did not impact upon viability.19 Furthermore, isolating EVs from plasma using SEC gave better quality RNAseq data for miRNAs as compared to EVs isolated using precipitation or UC.20 This emphasises the integrity of EVs isolated via SEC, allowing for their RNA cargo to remain safely inside the EVs until RNA extraction. As such, SEC is preferable to maintain target cell viability, maintain EV functionality and preserve EV RNA content.
Benefits of qEV isolation
Together, the purity, reproducibility and gentle nature of SEC have made it a growing and highly regarded choice for EV isolation.4,21 qEV columns are available in two formats to allow for the favouring of smaller (35 nm Series) and larger (70 nm Series) EVs, though both isolate a wide range of sizes to give good coverage of the spectrum of EVs within your sample. A variety of column sizes are also available to ensure optimal isolation from different starting volumes. qEV columns eliminate user-introduced variation that comes with ‘home-made’ SEC columns, allowing you to focus on your research without worrying about column quality. The use of the AFC automates the process of EV isolation to maximise reproducibility and even frees you to get on with something else in the lab whilst the PCV is being collected. Alongside the high adaptability of the AFC collection protocols, the variety of qEV columns allows for pure, reproducible isolation of EVs from your samples, regardless of starting material.









